Immune Signalling by Redox-Based Post-Translational Modifications
Redox changes are associated with the cell’s response to environmental stress cues. These changes are thought to contain information for down stream signaling processes that lead to transcription reprogramming. However, it remains largely unknown how cellular redox changes are sensed and translated into changes in gene transcription. Recent evidence indicates that cellular redox changes modify the status of inherently reactive cysteine residues. S-hydroxylation, S-sulphonation, S-nitrosylation, S-glutathionylation, and disulfide bonding are examples of reversible oxidative cysteine modifications that have profound effects on protein activity, conformation, interaction, and localization. Thus, these post-translational redox modifications are a powerful tool in cellular signaling (Spoel et al., 2010; Spoel and Loake, 2011).
As photosynthetic organisms, plants have inevitably evolved to cope with massive cellular redox changes. As a result plant cells contain a host of redox enzymes that are able to reduce differentially oxidized cysteine residues. Redox enzymes therefore provide the cell with the ability to fine tune post-translational redox modifications and employ them as signaling tools (Tada et al., 2008; Spoel et al., 2010; Spoel and Loake, 2011).
In our lab we aim to identify signaling proteins that are subject to regulatory redox modifications during plant stress responses. Moreover, we are interested in understanding the molecular mechanisms by which redox enzymes control the signaling properties of redox-sensitive cysteines.
For example, we previously addressed how in eukaryotes, bursts of reactive oxygen and nitrogen
species mediate cellular responses to the environment
by modifying cysteines of signaling proteins.
Cysteine reactivity toward nitric oxide (NO) leads to
formation of S-nitrosothiols (SNOs) that play important
roles in pathogenesis and immunity. However,
it remained poorly understood how SNOs are employed
as specific, reversible signaling cues. We showed that in plant immunity the oxidoreductase
Thioredoxin-h5 (TRXh5) reverses SNO modifications
by acting as a direct protein-SNO reductase (Kneeshaw et al., 2014).
Interestingly, TRXh5 restores a specific branch of SNO signalling that is different from the branch controlled by the indirect protein-SNO reductase, S-nitrosoglutathione Reductase (GSNOR1). Selective targeting of protien-SNO allowes TRXh5 to control signalling by the immune hormone salicylic acid. In summary, our data indicate that TRXh5 discriminates between protein-SNO substrates to provide previously unrecognized specificity and reversibility to protein-SNO signaling in plant immunity (Figure 1).
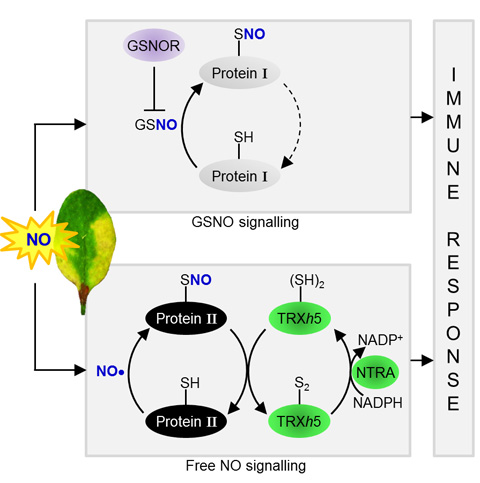
Figure 1. The protein-SNO Reductases GSNOR1 and TRXh5 Regulate Different Branches of Protein-SNO in Plant Immune
Signaling.
Two classes of proteins are shown. Class I proteins are S-nitrosylated by GSNO, the level of which is regulated by the indirect SNO reductase GSNOR1. Class II
proteins are S-nitrosylated by free NO, or other unknown intermediates and are denitrosylated by the TRXh5/NTRA system. Both pathways contribute to SA-dependent gene expression and immunity.

